For optimal use of this database, we recommend to use Google Chrome or Firefox browser.
Welcome to EXPath
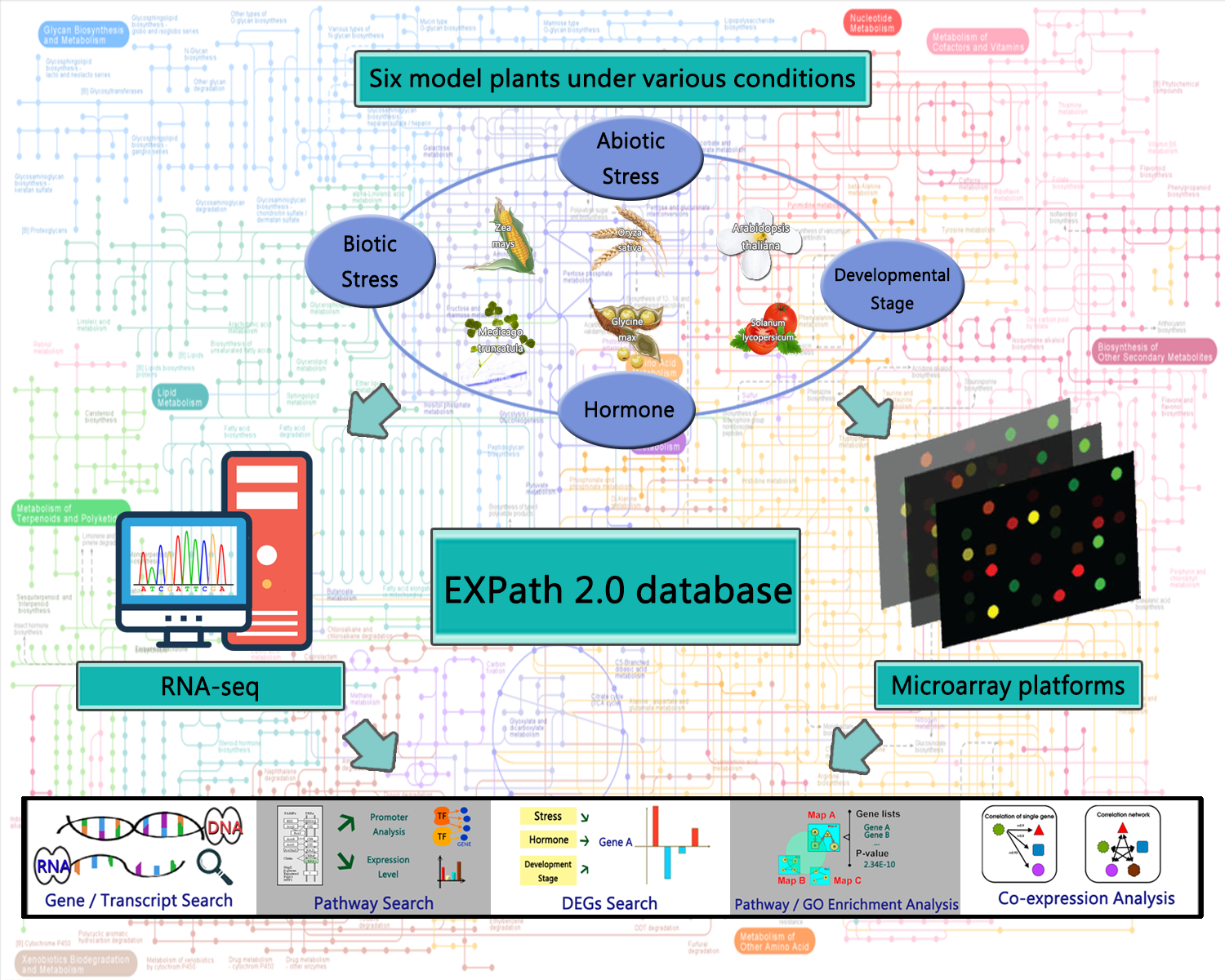
EXPath is a database resource that collects and utilizes expression profiles derived from microarray under various conditions to infer metabolic pathways for six model plants: Arabidopsis thaliana, Oryza sativa, Zea mays, Solanum lycopersicum, Glycine max, and Medicago truncatula. Five main functions in EXPath are as follows.
The current EXPath release (version 2.0) include:
★ Increase model crops from three to six
★ Establish
tissue-specific and organ-specific gene expression analysis
★ Construct distinct correlation networks for a group of genes under different conditions
★ Provide regulatory information of transcription factors involved in metabolic pathway or co-expressed gene group
Citation: Tseng, K.C., Li, G.Z., Hung, Y.C., Chow, C.N., Wu, N.Y., Chien, Y.Y., Zheng, H.Q., Lee, T.Y., Kuo, P.L., Chang, S.B. and Chang, W.C.* (2020) EXPath 2.0: An Updated Database for Integrating High-Throughput Gene Expression Data with Biological Pathways. Plant and Cell Physiology, 61, 1818-1827.
(yyyy/ mm)